ProteinStatsAI
Predict proteins statistics and properties using neural network.
If you are visiting here for the first time then I would recommend you to my earlier project DrugAI before reading any further. This project used SMILES signature of drug molecules to classify an organic molecule into different drug families.I have reused many functions and codes from this earlier project to this one. The main difference between this project and earlier project is, this is a regression problem whereas the previous one is a classifiction problem.
Similar to my earlier project I have used Bag of words (n-grams) [Link] for feature genration and then trained it in a nerual network. Here protein FASTA sequence is used as a feature. The dataset and its stats were obtained from pombase. Because the data were in two different files we have to first prepare, clean and save it in a CSV format before doing any machine learning. For data preparation I have used R program because its fast and easier to code.
Predicting targets
-
Mass
-
pI
-
Charge
-
NumResidues
-
CAI
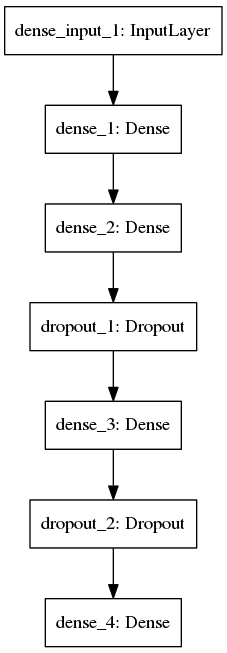
A simple multilayer perceptron is used for this study which was createad using keras module in python with a theanos backend. The FASTA sequence were converted to features and target values are normalized before feeding it to the neural network model. The model was trained for 15 epochs with a batch size of 200 for one cross validation (recommend to use 10). After training the project, it outputted a net validation loss of 0.0173422084428 and last validation loss of 0.0079427516942.
Project Github Page: https://github.com/Gananath/ProteinStatsAI